|

|

|

|

|

|

|
|
|
|
Infos
/ Organisation Meeting
|
 French Version
French Version
|
Reférences
Bibliographiques / References
|
Infos
Séquençages / Sequencing info
|
Contact
Participants / Reach Attendies
|
Crédits
/ Credits
|
|
|
|
For
the first time, scientists coming from different
perspectives have gathered themselves together to reflect on
the nature of the last common ancestor of all beings living
today. The Treille Foundation has enabled the organization
of a colloquium on this theme going beyond the usual
framework for scientific conferences. While numerous
participants presented several papers, some among them
improvised. Very quickly, the atmosphere of conviviality
appropriate to "Les Treilles" gave rise among all the
members of our little group (many of whom had never met
before) a sentiment of belonging to one community, at the
dawn of a new scientific adventure. Within a field (the
first step of life on earth) where the confrontation of
ideas (and now and then of personalities) could be
enlivened, the luminous atmosphere of Provence permitted the
discussion of even the more contradictory hypotheses in
complete serenity.
|
|
|
|

|
The first
objective of the colloquium was to assemble the
researchers appearing to come from different scientific
'tribes'. Each tribe has its own rites, in particular the
large conferences organized at regular intervals, its
journals of predilection and of great reputation. The
members of these different clans rarely mix , and only some
of the more audacious sometimes are accepted in two or three
different tribes. At "Les Treilles", at least five from
among these were represented: those researchers obsessed by
the problem of the origin of life, the
large international meeting on which will be held at
Orleans, that of the archaeo-microbiologists, specialists of
the third group of living beings on earth, the
archaebacteria or archea, that of the 'thermophilists' , who
study microbial life at very high temperature ( up to 110
°C) , that of the molecular evolutionists, who try to
retrieve the genealogy of all living beings (the universal
tree) by comparing the sequences of their macromolecules,
and finally the genomicists, a new tribe in formation, the
aim of which is the exhaustive analysis of genomes, rendered
possible thanks to large programs of systematic sequencing
of DNA.
|
|
Some
participants appear in none of the tribes mentioned, they
have been chosen by reason of their competence recognized in
a domain the importance of which appears crucial to confront
the problem of the last common ancestor. Some were
confronted for the first time with the great questions posed
by the evolution of life, and they were not any less
enthusiastic.
|
|
|
|
|
Most
of the researchers present were experimentalists,
they have behind them a long tradition of prudence, rigor,
going sometime to the point of opposition to all too
speculative ideas. In particular, reductionism of molecular
biology and the division which has been created since fifty
years ago between the 'pure and hard' biochemists and the
evolutionists are major obstacles in a field where the
imagination plays a determining role. From this point of
view, Christian De Duve had
played an irreplaceable role in the course of these days. A
Nobel prize winner, doyen of our colloquium, and recognized
for his work in cell biology, he has proved to have a
fertile imagination, playing a particularly active role in
our discussions. Our hopes are that the seminar at Treilles
will be the foundation of a new theme of wholly separate
research: research on LUCA (the
Last Universal Common Ancestor)
|
|
LUCA
was baptised at Treilles, yet we don't know if this name
imposed itself, but it was not unanimous between us. Other
terms have been proposed over the years: the
name progenote, put forward in 1977 by
Carl Woese (the creator of the
concept of archaebacteria) had its hour of glory, but it
seems on the decline. The notion of 'progenote' is in effect
associated with the conception of a particularly primitive
ancestor, much more simple than actual cells, but which does
not correspond any more with the thought of many
specialists. The majority vision today is more that of a
common ancestor resembling either bacteria or archaea, and
for some , that of a creature intermediate between
prokaryotes (cells without nuclei) and eukaryotes (cells
with nuclei).
|

|
|
|
The
term cenancestor
(from the Greek root cen. together) proposed by
Fitch in 1987, has the
favour of purists. For others, it presents the inconvenience
of being incomprehensible to ordinary mortals, and even to
the ordinary among biologists. The appelation 'Last Common
Ancestor' is more and more used in the literature.
José Castresana,
remarked during our seminar that it is also too vague. The
term 'last common ancestor' could be used (and is in effect)
for all groups of organisms.
|
|
|
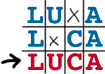
|
LUCA is a
compromise between LCA and LUA (last universal ancestor)
proposed at this colloquium by
Christos Ouzounis. It could be
popular with the media, since 'from
LUCA to LUCY' encapsulates or
replays the trajectory of the evolution of life. It defines
a sympathetic entity, one which seeded our planet- we are
all its descendants.s
|
|
Why
focus our attention on this key personage in our
history? Why a colloquium on this theme today? For a long
while research on the origins of life has been uniquely
devoted to understanding how the first molecules of life
appeared on our planet. Once they were present, the
consequences went of themselves( the rest followed). There
was no place for LUCA in these
researches. On the other hand, the cellular and molecular
biologists (with rare exceptions) did not want to waste
their time (very precious time) speculating on the ancient
unknowable, when the inexhaustible richness of model studies
offered themselves to them, those more real (in the form of
proteins and nucleic acids).
|
|
|
|
The
situation has much changed today, precisely thanks to
the progress of molecular biology. One has become aware , in
comparing the molecules of life of all organisms (from
bacteria to humans) that they display common points, a
fossilized record of their ancestral background. These are
the researches which have been conducive to the discovery of
the presence on earth of a group of prokaryotes, the
archaea, as distant from the bacteria as from the
eukaryotes.
|
|
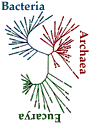
In
comparing the fundamental mechanisms of life in the
three Domains (bacteria, archaea, and eukaryotes) it then
becomes possible to define " the smallest common denominator
of the three": LUCA.
|
|
|
|
|
|
There
remains a division between the biologists loving
evolution, who also reconcile themselves with first origins
in going back to the past, and the pioneers of researches on
the origins of life, who try since the founder
experiment of Stanley
Miller in 1953 to start from point zero (the
primitive soup or dawn of stone) to advance towards the
present. The time has come to gather together all the
tribes.
|

|
|
Some questions debated at "Les
Treilles"
|
|
Is it possible to draw up an 'Identikit'
portrait of
LUCA
?
|
|

|
The
comparison of genes present in the three domains of
life is one of the favoured approaches for achieving
knowledge of LUCA. This
necessitates as a preliminary step the complete inventory of
genes present in each domain. This work will benefit from
the large number of genomes
which will be completely sequenced in the years to come.
A certain number of these have already been completed. The
colloquium at Treillles is being held within three months
after the publication of the complete sequence of the genome
of the yeast of the bakery (baker's yeast) , Saccharomyces
cerevisiae, and one month before the publication in Science
of the first
genome of an archaea, Methanococcus jannaschii.
Odile Ozier-Kalogeropoulos has
therefore presented the results obtained with the complete
analysis of S. cerevisiae, while Nikos
Kyrpides gave us a hint in advance of those obtained
in analysing the genome of Methanococcus jannaschii.
|
|
|
Christos
Ouzounis and
Nikos Kyrpides have presented a
first preliminary work comparing between genomes for
organisms belonging to all three domains. This work should
open on the inventory of proteins common to the three
domains, that is to say present similarities of sequence
which one could reasonably suppose to all be derived from
the same common ancestor (In technical terms, these proteins
are said to be homologs). These homologous proteins have a
strong chance of having been present in the
LUCA.
|
|
Right
now, the first results suggest a degree of complexity
unexpected in LUCA. That
organism would without doubt have already possessed many
thousands of genes (and therefore of different proteins). It
seems that all the large systems which permit the
maintenance and expression of genetic material were already
present, as were also many metabolic capacities.
José Castresana
contributed solid arguments to advance the iconoclastic idea
according to which the molecular mechanisms of oxygen
respiration were already present in
LUCA, despite an environment
apparently impoverished in oxygen! Was oxygen already
present on our planet in localized niches, or alternatively
did the mechanisms of oxidative respiration have another
function in the epoch of LUCA ?
|

|
|
|
One
question which is more general remains to be posed:
is the presence of the same gene in the three domains at the
present day always synonymous with the presence in
LUCA ? Was the transfer of
genes which distributed or could distribute throughout the
living world an invention which only appeared more lately in
one of the three domains? In particular,
Jim Brown and
Hervé Philippe have
shown that many protein phylogenies suggest the transfer of
a very large number of genes implicated in the metabolism of
bacteria to the eukaryotes, without doubt by the
intermediation of mitochondria, cellular organelles which
have evoloved in the eukaryote cells starting from ancient
endosymbiotic bacteria.
|

Communion around LUCA
|
|

|
To understand the origin
and evolution of real genomes with the object of going back
in time, it is equally necessary to understand the
mechanisms of their recent evolution. The comparative
analysis of genomes of organisms relative to the
evolutionary plan is very instructive from this point of
view.
Odile
Ozier-Kalogeropoulos has also compared a part of the
genome of the yeast Kluyveromyces lactis with that of S.
cerevisiae, while Renaud de
Rosa has compared the genome of two bacteria, the
coccibacillus Escherichia coli , of which more than 60% of
the sequence is known, and the pathogen Haemophilus
influenzae, of which the sequence has been completely
determined in the last year. These works put forward
evidence for very important differences in the rate of
evolution of one gene compared to another (and thus of one
protein to another) , of phenomena of rapid and massive loss
(relative to evolution) of certain genes, and finally a
large number of proteins called paralogs, that is to say
having diverged by duplication since the time of a common
ancestral gene.
|
|
The rapid
evolution of the sequence of certain proteins renders
difficult to determine evidence in support of their
relationship to one domain or to another. This could be in
part explained by the presence in the genomes completely
sequenced of many genes coding for proteins which have no
detectable homologs in the other two domains. In certain
cases, it is possible to find evidence for resemblance of
one domain to the other at the level of 3-dimensional
protein structure. Chris
Sanders has presented a work of structural analysis
which announces the putting in place of a systematic
strategy for identifying the very large number of genes and
functions possible in entire genome sequences. The number of
genes possessing homologs in the three domains (thus likely
to be present already in LUCA )
will also become augmented in years to come.
|
|
|
|
|
What root for the universal
tree
What can
be made of genes which are present only in two
domains? Were they already present in
LUCA and in this case, have
they been later (eventually) lost only in one of the three
domains, or have they really appeared in one branch common
to the two domains? the problem is complex; in effect
certain genes are present only in Bacteria or
Archaebacteria, others in archaebacteria and eukaryotes,
others again in Bacteria and the eukaryotes.
|

S. Miller & C. de Duve
|
|
If one
puts aside the problem of sampling, it will be a
priori easier to answer the question posed if one knows the
placement of the root of the universal tree which connects
the three Domains (between them). That is a question which
is very controversial. Many authors actually situate this
root in the Bacterial branch (the eukaryotes and
archaebacteria are in this case brother groups).
Jim Brown has presented some
results along this line, based on some universal trees
founded on proteins having diverged since the time of
duplication before the separation of the three domains.
|
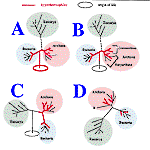
Location of LUCA and Hyperthermophiles
|
This result could explain the presence of numerous
characters of the "eukaryote" type within the archaea, as
well as protein phylogenies which associate archaea and
eukaryotes. It exists along with other phylogenies
associating Archaea and Bacteria. Jim
Brown interprets those in terms of the transfer of
genes from one Domain to the other.
|
|

|
The
validity of these results is
however contested by Patrick
Forterre and Hervé
Philippe, on the basis of studies on the variability
of the evolution of different positions of amino acids in
the same protein. According to them, there no longer remains
exploitable information permitting the rooting of trees
based on sequences having diverged so long ago. For
Hervé Philippe, aberrant
trees between proteins of three domains (for example those
which do not permit finding division into three of the
living world) are the norm, and it is the "coherent" trees
which demand to be explained. He suggests that the
appearance of "monophyletic" groups in a molecular tree
indicates an important switch in the structure/function of
the molecule studied.
|
|
It is important
to note that the position of the root of the
universal tree in the bacterial branch favors the idea by
which LUCA resembles actual
prokaryotes. Nevertheless, as long as the position of the
root is not known with certainty, the question remains open.
In particular , if the root is situated in the branch of the
eukaryotes, LUCA could in fact
resemble a cell of this type!
|
|
|
|
The eukaryote/prokaryote transition (or
vice-versa?)
For
Christian de Duve,
LUCA very much resembles a real
bacterium ( or archaeon). He presents an original scenario
which envisages the transition of prokaryotes to eukaryotes
in a quasi-solid milieu in which loss of the bacterial wall
is followed by the expansion of the cytoplasmic membrane to
permit phagocytosis. This expansion leads to the formation
of intracellular membrane networks characteristic of
eukaryote cells.
|
|
|

G. Ourisson & N. Glansdorff
|
Other
participants are in contrast in favor of a
LUCA in which the genome was
more of the eukaryote type. For
Nicolas Glansdorff, the operons
appeared slowly. perhaps in a lineage common to archaea and
to bacteria. For Rudiger Cerff,
the genome of LUCA was
fragmented into numerous chromosomes the genes of which
contained introns. Patrick
Forterre thinks that
LUCA was neither prokaryote nor
a eukaryote , but an organism of intermediate type , the
descendants of which have opted for opposite adaptive
strategies; adaptation towards miniaturization and the
maximum replication rate having led to prokaryotes, while
feeding on other organisms by predation led to the birth of
eukaryotes.
|
|
For
many participants,
LUCA was not an organism, but a
collection of diverse organisms exchanging their genes more
or less without constraint. In this respect was found debate
of the same type as that concerning the problem of the
African Eve. Without doubt a population geneticist would
have been needed to relate to our reflections. We regretted
the absence of Miroslav Radman,
prevented at the last minute from participating in our
colloquium. His lab studies the mechanisms of speciation at
a molecular level, and it seems that certain of these are
common to both prokaryotes and eukaryotes, suggesting that
the notions of species and species barrier existed already
during the epoch of LUCA ?
|
|
|
|

|
Another
essential question remains to be posed- what decisive
invention , appearing with
LUCA, gave it such a
predominance over its competitors of the epoch that only its
descendants today populate our planet?
|
|
|
|
Was
LUCA
a hyperthermophile?
Certain
authors think that life appeared at very high temperature
and that LUCA itself was a
hyperthermophile (an organism living between 80 and 110
°C). However, Stanley Miller
and Antonio Lazcano put
us on guard against the danger of extrapolating the supposed
conditions of life of LUCA to
that of the origins (of life). We have seen in effect that
LUCA was already a very
sophisticated organism, its appearance has therefore been
preceded by a long period of evolution.
Stanley Miller insists equally
on the instability of numerous prebiotic compounds at very
high temperature. Stanley Miller
and Patrick Forterre e
out the stress on the contradiction which exists between the
idea of a primordial evolution which be entirely produced at
high temperature (of the origin of
LUCA ) and the hypothesis of an
RNA world, taking into account the instability of this
molecule at temperatures in the neighbourhood of the boiling
point of water. Piero Cammarano
nevertheless presents some new facts on the evolution of
elongation factors (which participate in protein synthesis)
in favour of the ancientness (antiquity) of
hyperthermophilic bacteria. There again, the viability of
molecular phylogenetic analysis is at the centre of the
debate. Patrick Forterre
remarked that the results obtained with ribosomal RNA ,
which are generally interpreted in favour of the antiquity
of hyperthermophiles (their branches declare themselves by a
reduced length). could be tainted with error, given their
higher GC content, which has the effect of artificially
reducing the rate of evolution.
|
|
|
Nicolas
Glansdorff thinks that the
formation of operons could have been produced in response to
adaptation of prokaryotes to thermophilic conditions,
whereas Purificacion
Lopez-Garcia presented a new scenario which envisages
how an ancestral thermophile was modified in graduated steps
towards the bacteria and archaea. This scenario is based on
the study of enzymes which introduce supercoils into the DNA
molecule (gyrase and reverse gyrase) and on the hypothesis
of a precise geometry of this molecule necessary for its
function. This geometry will have been initially adapted to
temperatures in the region of 50 to 70 °C: the gyrase
and reverse gyrase will have then appeared in order to
permit he adaptation of organisms to lower and higher
temperatures respectively. In effect, the action of these
enzymes permits counteracting the effects of changes of
temperature on the DNA double helix (opening at high
temperature, compression at low temperature).
|
|
Whether
or not hyperthermophiles were near
LUCA, their study is a
particularly fascinating field of research.
Franck Robb presented some
experiments which contribute evidence on the mechanisms
permitting proteins of hyperthermophilic archaea to function
at temperatures in the neighbourhood of the boiling point of
water. One of the strategies utilized is the formation of a
network of ionic interactions at the surface of the protein.
He furthermore presented the first results showing that the
hyperthermophilic archaebacteria are particularly resistant
to ionizing radiation. These organisms should possess DNA
repair mechanisms of extremely good efficiency.
|
|
|
|
|
|
The age of
LUCA.
Some
months before holding our colloquium, an article
published in the American review Science rejuvenated
LUCA. It would have been two
billion years old, not 3.5 or 4 as one thought previously
according to examination of microfossils found in some
sedimentary rocks of this epoch which are still detectable.
Hervé Philippe has
subjected the facts of this work to a thorough critical
analysis from which he deduces (sets forth) that the date
advanced of 2 billion years rests on facts of very bad
quality. The protein phylogenetic trees used gave for the
most part aberrant results and the calibration of the
molecular clock is itself in question.
|

M. Fontecave & O. Ozier-Kalogeropoulos
|
|
|
|
Could we go back in time, beyond the
epoch where
LUCA
lived?
The
complexity of LUCA
suggests that a long period (at least in terms of
evolutionary changes) preceded its emergence. The first
steps of life on our planet had been the appearance and
assembly of the molecular constituents of life until the
appearance of he first cells. The genome of these first
cells was without doubt constituted by molecules of RNA and
not DNA. In contrast to DNA, RNA could in effect play at the
time the role of enzyme and of genetic material.
Stanley Miller and
Christian de Duve think that
the appearance of RNA was itself a late event , in effect,
this molecule does not seem to be able to be synthesized by
simple methods of prebiotic chemistry. RNA would have
therefore been preceded by molecules the exact nature of
which we will never know.
|
|
|

|
If until
LUCA
the first cells were RNA based , the road ought to
have been very long, always in terms of evolutionary
changes. Patrick Forterre
suggested dividing this evolution into several periods: the
two ages of the RNA world (before and after the invention by
RNA of a genuine system for synthesis of proteins) and the
first age of the DNA world (from the first DNA cell until
the emergence of LUCA. . How to
obtain information about these different stages? According
to him; the RNA virus and certain DNA viruses could be the
descendants of cellular organisms having lived before
LUCA. They would only have
survived the domination of the latter as parasites of its
descendants. The study of the molecular biology of viruses
could contribute information on the stages of evolution
before LUCA.
|
|
|
An
event essential for all this
evolution had been the invention of DNA. Today the synthesis
of DNA depends on a key enzyme, ribonucleotide reductase
(RNR), which makes the precursors of RNA (reduction of
ribose of RNA by elimination of oxygen to give a
deoxyribose). Marc Fontecave
has made the point of our knowledge on the 3 classes
of RNR known at present and on their distribution in the
living world. He presented the characterization by his team
of the first RNR of archaebacteria, in collaboration with
Franck Robb. This work has
permitted the demonstration for the first time that the
three classes of RNR derive from the same ancestral enzyme.
Marc Fontecave and
Franck Robb have taken
advantage of their common presence at Treilles to write up
an article reporting this discovery. The existence of an RNR
homologue in the three domains of life strengthens the idea
according to which the genome of
LUCA was already composed of
DNA.
|
|
|
To
establish the 'identikit'-portrait of predecessors of
LUCA,
Antonio Lazcano has proposed
using the paralog proteins the origin of which (by gene
duplication) is before the cell of
LUCA.
Arturo Becerro presented an
example in a work-in-progress of this strategy in the study
of the evolution of genes implicated in certain metabolic
pathways. Another example of comparative analysis of
metabolic pathways in the three domains was presented by
Nicolas Glansdorff. Again, the
problem is that of distinguishing duplications which may
have occurred before or after
LUCA , only the first can be
permitted in reconstituting the history of genes before
LUCA.
Bernard Labedan and
Renaud de Rosa have shown in
effect that recent duplications are numerous within the
bacteria and Odile
Ozier-Kalogeropoulos has reported results of the same
type in yeast.
|
|
|
|

P. Lopez Garcia, B. Labedan & R. deRosa
|
Bernard
Labedan summarized work in
collaboration with Monica Riley
on the protein
families of E. coli which suggested that many
ancestral proteins had already been formed from large
modules the shape and function of which were close to that
of genuine proteins. To rediscover the most ancient
homologies, we must without doubt make an analysis of
proteins in modular terms and perform structural analysis of
the type proposed by Chris
Sander.. The problem will then be to distinguish
between homology (shared structural heritage from a common
ancestor) and analogy (shared structure acquired by
convergent evolution). As a matter of fact, everyone agreed
on the necessity of combining genomic studies (comparison of
sequences) and structural facts which could be more
informative on the level of functions, for the purpose of
analysing evolution of he latter, which are the true targets
of natural selection.
|
|
|
Finally
the question poses itself: is it possible to
extrapolate from present-day metabolism to ancestral
metabolism? Guy Ourisson has
presented an attempt of this genre in the case of cellular
membranes which he considers as ought to have formed
(themselves) very early, perhaps before all other processes.
He has shown how the very wide variety of amphiphilic
membranes of the present day , often uncovered from
sediments in the form of their "molecular fossils", form an
evolutionary series. By retrograde analysis, it can be
deduced that the first membranes were probably formed by
phosphates of polyprenyl, which he has obtained by abiotic
reactions- and therefore probably prebiotic.
|
|
|
From his
perspective,
Christian de Duve
pointed out that present day metabolism up to
protometabolism preceded the RNA world (the world of
thioesters which he popularised in his book "Blueprints
of a Cell". He pitted himself against the absolutist
idea of a world of omnipotent RNA. According to him, all
present enzymes have not been preceded by the corresponding
ribozyme, and he would imagine a world of peptide catalysts
which were efficacious and very early diversified.
Stanley Miller, who has made
the point on the present state of research in the field of
origins of life in his introductory discussion, proposed in
conclusion a true programme of experimental research for
connections between prebiotic and contemporary metabolism
and advanced many new avenues.
|

P. Cammarano, P. Lopez Garcia, A. Lazecano & A. Becerra
|
|
In
conclusion,
Christian de Duve
emphasized the quality and intensity of the discussions. In
appearance, each had defended his point of view on the hotly
debated questions, but it was evident that the arguments
brought to bear by one and another will contribute to the
enrichment in the months to come of many reflections,
reposing of questions , or deepening (profound study) of
diverse hypotheses. The majority are going away only to meet
again for the editing of a series of articles to appear in a
special number of the international review "Journal of
Molecular Evolution" dedicated to this meeting. In the
opinion of many participants, it is one of the best
scientific meetings at which they have had the occasion to
participate (the setting of Treilles very much contributed
to this appreciation), and we all hope to renew this
experience in the years to come.
|
|
Several papers coming out of this exciting workshop
(including new collaboration initiated at "Les Treilles" )
have been published recently in a special issue of
"Journal of Molecular Evolution" Vol 49, Oct 1999.
|