
|
|















|
Genomes at IGM / LBMGE
|
Pyrococcus abyssi GE5 strain "Orsay" Genome description has been published in: |
|||
|
Sulfolobus solfataricus P2 Genome description has been published in: |
|||
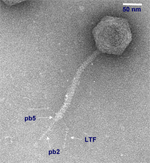 Bacteriophage T5 Bacteriophage T5st0 deletion mutant + wt insertion |
Bacteriophage T5 Sequence subcontracted to Integrated genomics This a joint project by: Transports Membranaires de Macromolécules, IBBMC Orsay France LBMGE IGM, Orsay France EAN IGM, Orsay France labs Unpublished GenBank accession numbers: AY692264 (st0 deletion mutant) & AY734508 wild type insertion |
||
 Kluyveromyces lactis Kluyveromyces lactis(image from wikimedia.org) |
Kluyveromyces lactis Sequence completed by Genoscope (http://www.genoscope.cns.fr/) Annotation of K. lactis is a joint project by IGM / Genolevures / Institut Pasteur Published in : Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisramé A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wésolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic Y, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL. Genome evolution in yeasts. Nature. 2004 Jul 1;430(6995):35-44. Zivanovic Y, Wincker P, Vacherie B, Bolotin-Fukuhara M, Fukuhara H. |
||
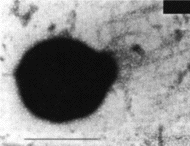 Thermococcus gammatolerans Thermococcus gammatolerans |
Thermococcus gammatolerans Free access Shotgun sequence by Genoscope This a project by: Laboratoire de Genomique des Archaea, IGM Orsay France Publication in Genome Biology on 06/11/09: |
||
 ΦX174 |
Viral Genomes : Microviridae in Sphagnum- dominated peatlands Free access Isolation / sequencing by UMR CNRS 6553 -ECOBIO, Universite de Rennes 1, Rennes, France Functional and Environmental Genomics platform (OSUR), Rennes, France This a project by: INSU-CNRS EC2CO program (VIRTOU) and grants from the University of Rennes/CNRS and from the observatoire des Sciences de l'Univers Rennes(OSUR) Publication in Frontiers in Microbiology: Diversity and comparative genomics of Microviridae in Sphagnum-dominated peatlands. Quaiser A, Dufresne A, Ballaud F, Roux S, Zivanovic Y, Colombet J, Sime-Ngando T and Francez A-J (2015) Front. Microbiol. 6:375. | http://dx.doi.org/10.3389/fmicb.2015.00375 |
||
 Cruciviridae |
Viral Genomes : Cruciviruses: chimeric viruses in Sphagnum- dominated peatlands Free access Isolation / sequencing by UMR CNRS 6553 -ECOBIO, Universite de Rennes 1, Rennes, France Functional and Environmental Genomics platform (OSUR), Rennes, France This a project by: INSU-CNRS EC2CO program (VIRTOU) and grants from the University of Rennes/CNRS and from the observatoire des Sciences de l'Univers Rennes(OSUR) Publication Quaiser, A., Krupovic, M., Dufresne, A., Francez, A. J., & Roux, S. (2016). Diversity and comparative genomics of chimeric viruses in Sphagnum-dominated peatlands. Virus evolution, 2(2), vew025. | https://doi.org/10.1093/ve/vew025 |
||
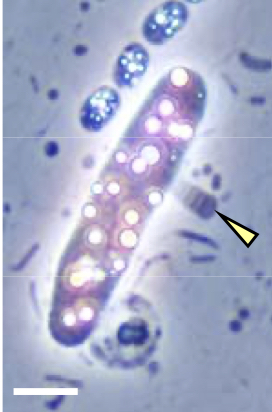 Vampirococcus lugosii Vampirococcus lugosii |
Vampirococcus lugosii Free access Isolation / amplification / sequencing by Ecologie Systématique Evolution, CNRS, Université Paris-Saclay, AgroParisTech, Orsay, France. Institut de Biologie Intégrative de la Cellule, CNRS, Université Paris-Saclay, Orsay, France This a project by: ESE, CNRS, Université Paris-Saclay, AgroParisTech, Orsay, France. I2BC, CNRS, Université Paris-Saclay, Orsay, France Publication NATURE COMMUNICATIONS | (2021)12:2454 | https://doi.org/10.1038/s41467-021-22762-4 |
||
 Deinococcus radiodurans R1 ATCC 13939 |
Deinococcus radiodurans R1 ATCC 13939 Free access Isolation / amplification / sequencing by Universite Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198 Gif-sur-Yvette, France. Publication Cells | (2021) Sep 25;10(10):2536. | https://doi.org/10.3390/cells10102536 |
||
 Candida glabrata CBS138 |
Nakaseomyces genomes backup database Free access Maintained at Universite Paris-Saclay, CEA, CNRS, Institut Diversité, Écologie et Évolution du Vivant (IDEEV), 91198 Gif-sur-Yvette, France. This is a custom database of publicly available Nakaseomyces genomes/scaffolds for the benefit of local users. Publications GenBank accession numbers: |
||
Pyrococcus abyssi is an hyperthermophilic euryarcheaon thriving in deep-sea hydrothermal vents , growing optimally at 96°C in strict anaerobic conditions and metabolizing sulfur. It has been sampled in the North Fiji basin (SW Pacific) at 2000 mt depth in 1989.
Sulfolobus solfatricus is an aerobic crenarchaeon that grows optimally at 80°C and pH 2-4 metabolizing sulfur which growths in volcanic solfataras. It is the most widely studied organism of the crenarchaeal branch of the Archaea
Bacteriophage T5 is a lytic, linear, double stranded "nicked" DNA phage belonging to Siphoviridae family. The phagic infection is a two step mechanism, in which the first-step transfer segment allows expression of pre-early genes necessary for complete DNA transfer and initiation of the lytic cycle. We have sequenced the deletion mutant st0, a widely used laboratory strain for primary sequence determination. Subsequently, full wild type genome has been reconstructed by incorporating the sequence of the wild type insertion from HER28 strain (obtained from Reference Center For Bacterial Viruses, Laval University, Quebec, Canada)
Kluyveromyces lactis, a yeast commonly used in genetic studies, cheese and yogurt production, and could potentially be used to produce drug compounds.
A large scale comparative genomic study as been undertaken, leading to sequencing and comparison of 3 other hemiascomycetous yeasts (Candida glabrata, Debaryomyces hansenii, and Yarrowia lipolytica.)
Thermococcus gammatolerans, was isolated from samples collected from hydrothermal chimnneys. This organism withstands 5 kGy irradiation without any detectable lethality and thus it is one of the most radioresistant organisms known amongst the Archaea. Complete sequencing, assembly and annotation was performed at LGA, Orsay. Complete proteome analysis performed in collaboration with CEA, DSV, IBEB Laboratoire de Biochimie des Systèmes Perturbés, Bagnols-sur-Cèze, F-30207, France. Transcriptonal response to Cadmium and gamma-irradiation analysis performed at LGA.
LBMGE has been directly involved in genome sequencing of these two hyperthermophilic archeons during the past years.
Both strains are widely studied model microorganisms in various fields of investigation such as studies on adaptation to extreme temperatures and pH growth conditions, aerobic and anaerobic sulfur metabolism analysis, molecular deciphering of DNA replication mecanism in archaea, phylogeny and rooting of the tree of life, comparative genomics of closely related strains and many others.
This page provides direct links to genomic annotation databases for P. abyssi and S. solfataricus, bacteriophage T5 and T gammatolerans
All databases on this site are maintained by Y. Zivanovic.
Some references related to these projects:
P. abyssi:
- Lopez, P., Philippe, H, Myllykallio, H. and Forterre, P.
Identification Of Putative Chromosomal Origins Of Replication In Archaea.
Mol. Microbiol. 32, 883-886 (1999) - H. Myllykallio, P. Lopez, P. López-García, R. Heilig, W. Saurin, Y. Zivanovic, H. Philippe, and P. Forterre
Bacterial Mode Of Replication With An Eukaryotic-Like Machinery In A Hyperthermophilic Archaeon
Science, 288, 2212-2215 (2000) - O. Matte-Tailliez, Y. Zivanovic and P. Forterre
Mining archaeal proteomes for eukaryotic proteins with novel functions: the PACE case
Trends Genet. 16, 533,536 (2000) link on this site : PACE Proteins - Zivanovic, Y., Myllykallio, H. and Forterre, P.
Bacterial genomic reorganization upon DNA replication.
Science 292, 803a (2001) - Zivanovic Y., Lopez P, Philippe P and Forterre P
Pyrococcus genome comparison evidences chromosome shuffling-driven evolution.
Nucleic Acids Research, 30, 2656-62 (2002) - Cohen G., Barbe V., Flament D., Galperin M., Heilig R., Lecompte O., Poch O., Prieur D., QuŽrellou J., Ripp R., Thierry J., Van J. der Oost, Weissenbach J., Zivanovic Y. and Forterre P.
An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi
Mol. Microbiol. in Press (2002)
S. solfataricus:
- She, Q., Singh, R.K., Confalonieri, F., Zivanovic, Y., Gordon, P., Allard, G., Awayez, M.J., Chan-Weiher, C-Y., Clausen, I.G., Curtis, B., De Moors, A., Erauso, G., Gordon, P.M.K., Heikamp de Jong, I., Jeffries, A., Kozera, C.J., Medina, N., Peng, X., Phan, H., Redder, P., Schenk, M.E., Theriault, C., Tolstrup, N., Charlebois, R.L.M., Doolittle, W.F., Duguet, M., Gaasterland, T., Garrett, R.A., Ragan, M., Sensen, C.W. and Van der Oost, J.
The genome sequence of the crenarchaeon Sulfolobus solfataricus P2.
Proc. Natl. Acad. Sci. USA 98, 7835-7840. (2001) - Forterre, P., Confalonieri, F. and Knapp, S.
Identification Of The Gene Encoding Archaeal-Specific Dna Binding Proteins Of The Sac10b Family
Mol. Microbiol. 32, 669-670 (1999) - R. Charlebois, R. Singh, C.C.-Y. Chan-Weilher, G. Allard, C. Chow, F.,Confalonieri, B. Curtis, M. Duguet, D. Faguy, T. Gaasterland, R. Garrett, A.,Jeffries, C. Kozera, N. Kushwala, E. Lafleur, N. Medina, X. Peng, S.,Penny,Q. She,A. Stjohn, J. Van Der Oost, F. Young, Y. Zivanovic, W.,Doolittle, M. Ragan & C. Sensen.
Gene Content and Organization Of A 281 Kbp Contig From The Genome Of The Extremely Thermophilic Archaeon Sulfolobus Solfataricus P2.
Genome 43, 116-36 (1999) - Q She, F Confalonieri, Y Zivanovic, N Medina, A Billault, M J. Awayez, H P, Thi-Hgoc, B-T T Pham, J Van Der Oost, M Duguet, and R Garrett
A Bac Library and Paired-PCR Approach To Mapping and Completing The Genome Sequence Of Sulfolobus Solfataricus P2: An EC Perspective
DNA Seq., 11(3-4),183-92. (2000) - Brügger K., Redder P., She Q., Confalonieri F., Zivanovic Y. and Garrett R. A.
Mobile elements in archaeal genomes
FEMS Microb. Lett., 206, 131-141 (2002)
Kluyveromyces lactis:
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisramé A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wésolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic Y, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL.
Genome evolution in yeasts.
Nature. 2004 Jul 1;430(6995):35-44. - Zivanovic Y, Wincker P, Vacherie B, Bolotin-Fukuhara M, Fukuhara H.
Complete nucleotide sequence of the mitochondrial DNA from Kluyveromyces lactis.
FEMS Yeast Res. 2005 Feb;5(4-5):315-22.
- Recovery of ionizing-radiation damage after high doses of gamma ray in the hyperthermophilic archaeon Thermococcus gammatolerans.
Tapias A, Leplat C, Confalonieri F.
Extremophiles. 2009 - Genome-wide transcriptional response of the archaeon Thermococcus gammatolerans to Cadmium
A Lagorce, A Fourcans, M Dutertre, B Bouissiere, P Forterre, X Gidrol, Y Zivanovic And F Confalonieri
PLoS One. 2012;7(7):e41935. doi: 10.1371/journal.pone.0041935. Epub 2012 Jul 27. PMID: 22848664; PMCID: PMC3407056. - Genome analysis and genome-scale proteomics of Thermococcus gammatolerans, the most radioresistant organism known amongst the Archaea.
Y. Zivanovic, J. Armengaud, A. Lagorce, C. Leplat, P Guerin, M. Dutertre, V. Anthouard, P. Forterre, P. Winker, and F. Confalonieri
Genome Biol. 2009;10(6):R70. doi: 10.1186/gb-2009-10-6-r70. Epub 2009 Jun 26. PMID: 19558674; PMCID: PMC2718504.

